1. Defining the role of novel driver genes in therapy resistant breast cancer and their interactions with the tumour microenvironment
A major challenge for the translation of genomic data generated from next-generation sequencing studies into patient benefit and precision medicine is distinguishing ‘driver’ alterations (provide a selective advantage to the cell harbouring its mutation at a given point in time) from ‘passenger’ (those that have no biological significance on the cell harbouring its mutation at a given point in time). The majority of the novel mutations observed in the common types of breast cancer occur at relatively low frequency, which makes distinguishing drivers from passengers difficult. Nevertheless, recent massive parallel sequencing efforts have revealed a diverse set of low frequency genetic events that are potentially targetable highlighting the need for functional characterisation of the long tail of mutations within heterogeneous tumours. Within the context of poor prognosis breast cancers, identifying molecular drivers of disease progression is of paramount importance. In particular, given the recent advances in disease monitoring by circulating free DNA, identification of actionable alterations would increase therapeutic options.
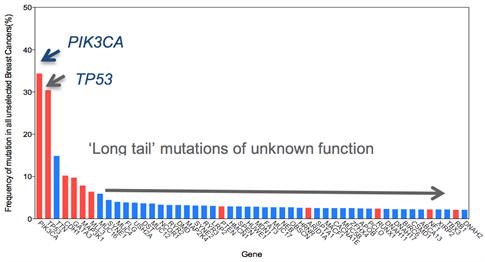
Figure 1: Frequency of mutations in unselected breast cancer. Red highlights known driver genes. Note many of these well characterised driver genes are present at low frequencies.
The Functional Genomics Team has a track record in the functional analysis of driver mutations in breast cancer (PPM1Dand CCNE1amplifications, CDK12fusions and SF3B1mutations in different subtypes of breast cancer (Natrajan et al Clin Cancer Res 2009, Natrajan et al Breast Cancer Res 2012, Natrajan et al J Pathol 2014, Maguire at al J Pathol 2015, Naidoo et al Mol Cancer Ther 2018). Employing bioinformatics mining, and functional assessment we seek to define the role of these alterations in disease progression and explore new avenues for therapeutic intervention.
Defining the interplay with microenvironmental pressures
In particular, to define the role of novel driver mutations we have developed a a 3-dimensional cell culture functional genomics screening platform to model more aggressive tumours in the form of cancer cell line spheroids to identify novel loss of function alterations that drive progression (Maguire and Peck et al J Pathol 2016 and Morrison et al JoVe 2016). Using this platform, we have identified novel tumour suppressor genes involved in breast cancer progression of therapy resistant disease. In particular, these alterations impart a selective advantage only under 3D conditions, indicating their driver capacity is dependent on particular selective pressures such as oxygen and nutrient stress and furthermore are associated with therapy resistant forms of breast cancer. We aim to use further investigate the role of these driver genes using in vitroand in vivopatient derived models as well as GEMM models to dissect their role in metastatic progression. In addition, we aim to use high-throughput CRISPR, siRNA and drug functional genomic screens to identify vulnerabilities in these tumours harbouring these alterations and identify therapeutic approaches that may be useful in these patients.
Current areas of investigation include aberrations in chromatin remodelling complexes.
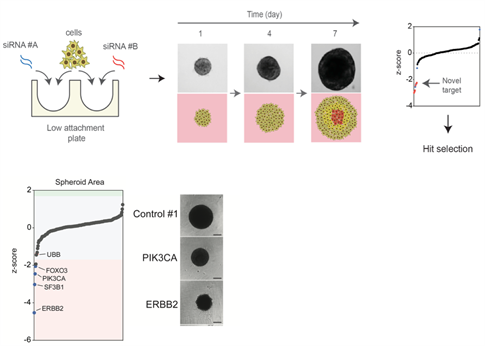
Figure 2: Identification of novel functional genomics pipeline for functional assessment of driver alterations of aggressive disease. Cells are reverse transfected and seeded into low attachment plates to allow spheroid formation. Growth is monitored over time to identify genes that modulate tumour growth in a 3D dependent manner. Example of a proof of principle 3D siRNA screen output identifying ERBB2 and PIK3CA silencing inhibits ERBB2 amplified and PIK3CA mutant cell growth in 3D.
2. The role of spliceosomal reprogramming in aggressive ER+ breast cancer
We have identified a new role on tumour progression in aggressive ER+ breast cancer for mutations in components of the spliceosome that lead to global splicing alterations increasing heterogeneity, and can be therapeutically targeted through inhibition of the spliceosome itself (Maguire et al J Pathol 2015, Read et al Endocr Relat Cancer 2018). We are further dissecting the role of spliceosomal alterations in ER+ breast cancer through targeted and genome wide functional genomic screens and exploring the functional consequences of aberrant splicing in SF3B1mutant cells. We are also exploring whether alternative spliced isoforms can serve as novel therapeutic biomarkers in treatment resistant breast cancers.
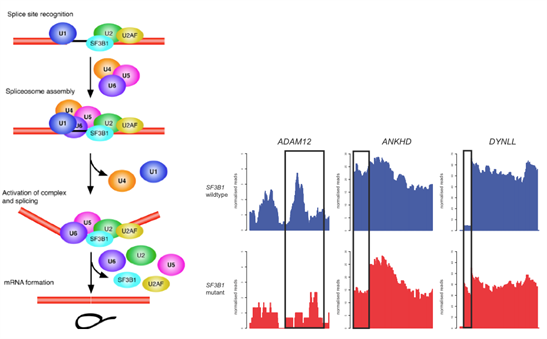
Figure 3: Consequences of aberrant splicing in SF3B1mutant breast cancer.
Cartoon depiction of spliceosome formationdepicting the role of SF3B1 in 3’ splice site recognition. Normalised RNA-sequencing plots depicting splicing alterations identified in primary breast cancers SF3B1mutant (red and wild-type (blue).
3. Tumour Heterogeneity and its role in treatment resistance
Single-cell profiling of patient tumors and of in vivomodels are revealing that many cancers are constituted of genetically and phenotypically distinct clones that can interact with one another. In particular, we have shown through image analysis and ecological statistical approaches that heterogeneity between different cell populations in breast cancer governs breast cancer clinical outcome (Natrajan et al PLoS Medicine 2016). We aim to study how the clonal dynamics within tumours are altered by treatment with standard and experimental treatments by integrating image analysis, in situ analysis and high throughput sequencing approaches including single cell sequencing approaches. Through this we aim to identify new genetic and phenotypic biomarkers of treatment resistance.
We also use in vitro and in vivomodels coupled with computational modelling to study the clonal dynamics within tumours and responses to therapy to identify molecular mechanisms of therapy resistance and possible new targets.
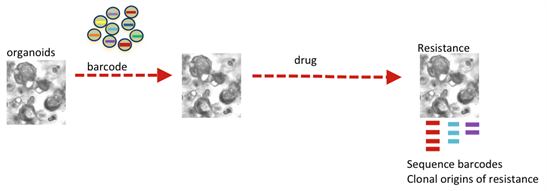
Figure 4: Modelling clonal dynamics over time using molecular barcoding.
4. Development of novel in vivo and in vitro models of breast cancer progression
All our work is underpinned by more relevant models (3D) and in vivomodels derived from primary patient material. In particular, we have implemented the novel MIND (mouse intraductal model) to study breast cancer progression. This model more accurately recapitulates disease phenotype and can also model in situto invasive disease. We aim to develop new patient derived models using the MIND approach to further understand disease progression.
In addition, we have characterised the in vitro MCF10A progression series (Maguire and Peck J Pathol 2016) which provides an excellent isogenic platform to dissect the role of alterations on different stages of disease progression.
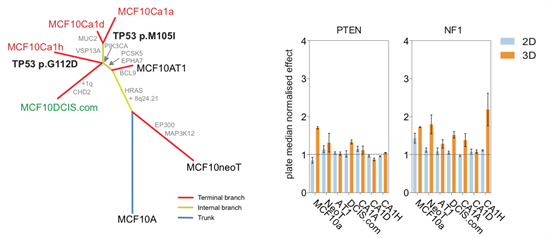
Figure 5: Evolutionary tree of MCF10 progression seriesidentifies acquired alterations associated with progression from non-malignant (MCF10A, MCF10neoT, MCF10AT1) to DCIS (MCF10DCIS.com) through to invasive disease (MCFCa1a, MCF10Ca1d and MCF10Ca1h). Viability plots showing growth advantage only occurs in 3D when known tumour suppressor genes are silenced in the MCF10 progression series, highlighting roles in early (PTEN) and later stage (NF1) progression.