Structure of a critical protein for cell division is imaged
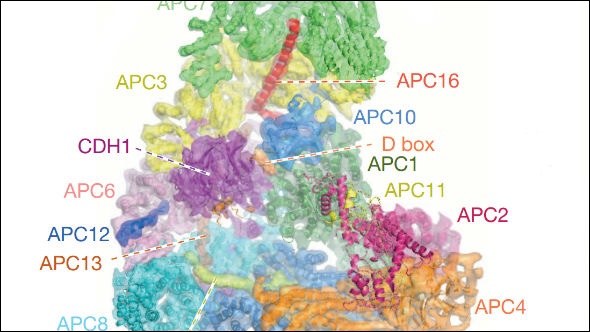
Professor David Barford, former Professor of Molecular Biology at the ICR, revealed the structure of one of the most important and complicated proteins in cell division – and cancer.
A team at the ICR in London and the Medical Research Council Laboratory of Molecular Biology in Cambridge produced the first detailed images of the anaphase-promoting complex (APC/C).
The APC/C performs a wide range of vital tasks associated with mitosis, which is used in cell division by all animals and plants, and is the process by which a cell copies its chromosomes and pulls them apart into two separate cells.
Images of the gigantic protein in unprecedented detail will transform scientists’ understanding of exactly how cells copy their chromosomes and divide, and could reveal binding sites for future cancer drugs.
The research, which was published in Nature, used advanced electronic microscopy and imaging software to map the APC/C in extremely high detail, with a resolution of less than a billionth of a metre.
The researchers were able to spot the individual building blocks of APC/C – helixes and folded sheets of amino acids that compose the tri-dimensional structure of a protein.
Researchers identified 20 subunits in APC/C, which interact with each other and with the cell environment during the cell cycle. They allow the complex to control a range of mitotic processes including the initiation of DNA replication, the segregation of chromosomes along protein ‘rails’ called spindles, and the ultimate splitting of one cell into two, called cytokinesis. Each of these processes is a potential target for disruption for potential new cancer drugs designed to stop replication of cancer cells.
The study was funded by Cancer Research UK.
References
Chang L. et al., (2014) Molecular architecture and mechanism of the anaphase-promoting complex, Nature 513, 388-393, doi:10.1038/nature13543
Enzalutamide improves patient survival in men with prostate cancer before chemotherapy
Professor Johann de Bono at the ICR and The Royal Marsden helped lead a phase III clinical trial demonstrating that the prostate cancer drug enzalutamide improves patient survival in men with advanced prostate cancer who had not received chemotherapy (image: Mateus Crespo/Professor Johann de Bono, the ICR)
In the trial, researchers gave enzalutamide to more than 800 men with castration-resistant prostate cancer who no longer responded to standard hormone-suppressing drugs but who had not yet received chemotherapy. The study, published in the New England Journal of Medicine, found that enzalutamide extended the time before tumours started progressing and improved overall survival. The drug acts by blocking androgen receptors - halting the growth of prostate tumours, which rely on androgens to grow.
After 12 months, 65% of men treated with enzalutamide had no detectable tumour progression, compared with only 14% of men receiving the placebo. The drug also delayed the need for treatment with chemotherapy by an average of 17 months.
Enzalutamide has already been approved by NICE for use in men with advanced prostate cancer who have already received chemotherapy. But the new study suggests that the drug could benefit men with prostate cancer earlier in the course of their disease, without them having to undergo toxic chemotherapy first, and could expand the treatment options available at this stage of treatment.
Advanced prostate cancer has historically been extremely difficult to treat, but following a coordinated effort to bring new drugs to patients, there has been dramatic progress in treatment in recent years. Drugs like enzalutamide and abiraterione, which was discovered at the ICR, are helping to extend men’s lives and improving their quality of life.
References
Beer T.M. et al, (2014) Enzalutamide in Metastatic Prostate Cancer before Chemotherapy, The New England Journal of Medicine, 371, 424-433, doi:10.1056/NEJMoa1405095
Genetic flaw links childhood brain cancer to stone man syndrome
ICR researchers led by Dr Chris Jones discovered a remarkable link between a type of childhood brain cancer and an inherited disorder known as stone man syndrome.
They identified a genetic flaw common to both the brain cancer diffuse intrinsic pontine glioma (DIPG) and fibrodysplasia ossificans progressiva (FOP) – a rare syndrome in which muscle tissue gradually turns to bone. Drugs that target the genetic flaw could offer hope as a treatment for both devastating diseases.
DIPG is a devastating form of cancer, affecting about 20-30 children per year in the UK. Survival is almost always less than a year and with no available treatment currently available, the disease is invariably fatal.
The researchers examined biopsy samples from children with DIPG and identified the genetic defect in a quarter of them. The defect appears to trigger DIPG when it occurs within the brain, and while it hasn’t been linked to any other cancer, it is known to cause FOP when it is inherited.
But the researchers hope that their study, which was funded by Cancer Research UK and published in the journal Nature Genetics, could pave the way for new cancer therapies that could offer hope to children with DIPG for the first time. There are already drugs under development to treat FOP, targeted at the shared genetic defect, and these belong to a drug class that has also been used to treat other forms of cancer. There is hope that these drugs or others like them could be effective treatments for both diseases.
References
Taylor K. et al., (2014) Recurrent activating ACVR1 mutations in diffuse intrinsic pontine glioma, Nature Genetics 46, 457- 461, doi:10.1038/ng.2925
Mutations in leukaemia gene linked to new childhood growth disorder
A study led by Professor Nazneen Rahman at the ICR discovered that mutations in a gene associated with leukaemia cause a newly described condition that affects growth and intellectual development in children.
The study identified mutations in the DNA methyltransferase gene, DNMT3A, in 13 children.
All the children were taller than usual for their age, shared similar facial features and had intellectual disabilities. The mutations were not present in their parents, nor in 1,000 controls from the UK population.
The new condition has been called ‘DNMT3A overgrowth syndrome’.
The DNMT3A gene is crucial for development because it adds the ‘methylation’ marks to DNA that determine where and when genes are active.
Intriguingly, DNMT3A mutations are already known to occur in certain types of leukaemia. The mutations that occur in leukaemia are different from those in DNMT3A overgrowth syndrome and there is no evidence that children with DNMT3A mutations are at increased risk of cancer.
Researchers at the ICR, with colleagues at St George’s, University of London, The Royal Marsden NHS Foundation Trust, and genetics centres across Europe and the US, identified the mutations after analysing the genomes of 152 children with overgrowth disorders and their parents.
The discovery is of immediate value to the families of affected children in providing a reason for why their child has had problems. And because the mutations have arisen in the child and have not been inherited from either parent, the risk of another child in the family being similarly affected is very low.
The research, published in the journalNature Genetics, is a part of the Childhood Overgrowth Study, which is funded by the Wellcome Trust, and aims to identify causes of developmental disorders that include increased growth in childhood.
References
Tatton-Brown K et al., (2014) Mutations in the DNA methyltransferase gene DNMT3A cause an overgrowth syndrome with intellectual disability, Nature Genetics 46, 385-388, doi:10.1038/ng.2917
Cullin-5 enzyme plays role in response to key class of cancer drugs
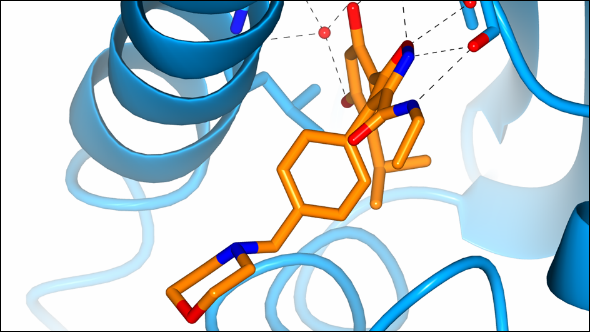
Professor Paul Workman, Chief Executive of the ICR, discovered with his team new roles for a molecule that responds to the activity of an important class of cancer drugs.
The findings point to a possible test that could identify patients who would be most responsive to the drugs – called Hsp90 inhibitors - and who might develop resistance.
Professor Workman was responsible with his team for driving the science underpinning Hsp90 inhibitors, and for discovering one of the leading drugs in the class, AUY922.
The new study, led by Professor Workman, also suggests new approaches to make Hsp90 inhibitors more effective or discover other types of cancer drug.
The research, published in the journal Proceedings of the National Academy of Science (PNAS),showed that an enzyme called Cullin-5 (CUL5) works to clean up proteins that tell cancer cells to keep dividing, and then disposes of them.
CUL5 seems to help determine the response to Hsp90 inhibitors, which work by blocking the molecule heat shock protein 90 - a chaperone for a number of other proteins related to cancer growth.
In the study, scientists found that when cancer cells were treated with Hsp90 inhibitors, CUL5 immediately stepped in to ‘bin’ the proteins that were telling cancer cells to keep dividing.
CUL5 also seemed to help pull the ‘dividing signal’ proteins away from the protective shelter of HspP90, and label them with a tag sending them to the cellular dustbin.
Silencing CUL5 led some cancer cells to become less sensitive to Hsp90 inhibitors, suggesting that patients whose cancers express lower levels of the enzyme could be at risk of becoming resistant to Hsp90 inhibitors.
Hsp90 inhibitors are a highly promising new cancer treatment, so biomarkers that could predict a patient’s response to treatment could be of significant clinical benefit.
References
Samant. R.S. et al., (2014) E3 ubiquitin ligase Cullin-5 modulates multiple molecular and cellular responses to heat shock protein 90 inhibition in human cancer cell, Proceedings of the National Academy of Science, 111, 18, 6834–6839, doi: 10.1073/pnas.1322412111
‘Non-stick’ gene can prevent breast cancer spread
A team at the ICR led by Professor Clare Isacke found that a gene can prevent the spread of breast cancer by stopping tumour cells from becoming ‘sticky’.
Drugs to mimic the effect of the gene could help prevent the cancer cells from metastasising – spreading cancer to other parts of the body – by stopping them from developing stickiness.
The study, which was published in the journal Cancer Discovery, used the powerful technique of RNAi screening to switch off more than 1000 breast cancer genes.
Researchers then analysed secondary tumours which developed in the lungs of mice to assess the effect of knocking out specific genes on the behaviour of cancers.
The researchers identified several genes which contributed to the formation of secondary tumours when switched off, one of which was the ST6GalNAc2 gene. The gene prevents cancer cells from attaching a protein called galectin-3 to their surface which gives them their stickiness.
Some women with breast cancer may be at higher risk of their cancer spreading to new parts of the body because of the silencing of this gene. The researchers found that ST6GalNAc2 gene activity was particularly low in a subtype of breast cancer patients who have oestrogen receptor negative (ER-) breast cancer.
The results suggest that administering existing, well-tolerated drugs that target galectin-3 activity to women with ER- breast cancer could potentially slow the spread of their cancer. Clinical trials have already shown these drugs to be safe and the findings of this study could open up a new treatment approach for the prevention of cancer spread.
References
Murugaesu N. et al., (2014) An In Vivo Functional Screen Identifies ST6GalNAc2 Sialyltransferase as a Breast Cancer Metastasis Suppressor, Cancer Discovery, 4, 275-277, doi: 10.1158/2159-8290.CD-13-028
Protein complex helps cancer cells with multiple centrosomes to divide
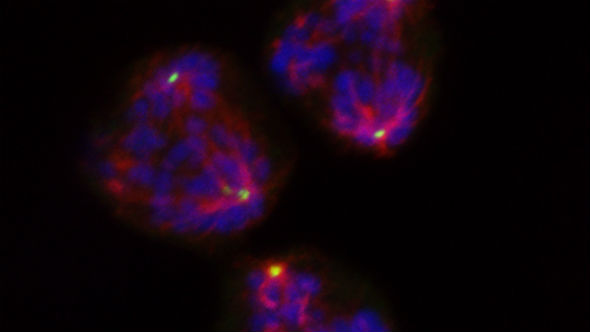
Cancer cells use a protein complex to divide even when they have multiple centrosomes – which normally come only in pairs. But cell division using multiple centrosomes represents a fundamental weakness in cancer cells which could be targeted by drugs, an exciting study led by Dr Spiros Linardopoulos in our Division of Breast Cancer Research found.
The study, published in Nature Communications, looked in great detail at how cancer cells gather together the tiny centrosomes at the time at which the cells divide. Centrosomes play a crucial part in the hugely complex process of cell division, acting as marshals for the rail-like spindles along which DNA is pulled apart when one cell becomes two.
Cancer cells are highly genetically unstable, and over the past years, researchers have realised that they often accumulate many more centrosomes than in normal, healthy cells.
These extra centrosomes should play havoc with the cell division process and kill cancer cells by pulling DNA in many different directions at once – but cancer cells have developed ways to gather them into clusters to keep them out of the way.
The study, which was carried out in breast cancer cells, showed the vital importance of a super-protein called APC/C in this process of centrosome clustering. It showed that the APC/C could be a target for drugs that scramble cancer’s cell division machinery, killing them while leaving most healthy cells intact.
The study – which was funded by Breakthrough Breast Cancer – showed that APC/C’s function in centrosome clustering was disrupted by small chemical molecules , proving the principle that the super-protein is druggable. The study could ultimately lead to the development of new drugs which target this previously unexploited weakness in cancer.
APC/C, or the anaphase-promoting complex, was also the focus of another one of our top 10 discoveries of the year, – that study, led by researchers in our Division of Structural Biology, mapped the protein in unprecedented detail.
References
Drosopoulos K et al, (2014). APC/C is an essential regulator of centrosome clustering, Nature Communications, 5: 3686. doi:10.1038/ncomms4686.
Imaging technique assesses treatment in advanced prostate cancer
Dr Simon Robinson and colleagues from the ICR developed a way to test how effective drugs are likely to be against prostate cancer that has spread to the bone.
Their mouse model closely resembles how the disease develops in human bone and could help researchers to accelerate the progress of new drugs into the clinic.
Dr Robinson and colleagues combined multiple imaging techniques to show clearly in mice how tumours and the surrounding bone were affected by a drug called cabozantinib.
Prostate cancer affects around 41,700 men in the UK each year. Over 80 per cent of men will survive the disease for at least five years, but prostate cancer that has spread to the bone is much harder to treat, and there has been a need to develop good models of the disease in this setting.
The drug cabozantinib blocks two proteins called MET and VEGFR2, which are important for normal cellular processes, but also help cancer cells to spread and invade parts of the body such as the skeleton.
Researchers wanted to use their new mouse model to investigate whether blocking the activity of these proteins could slow or stop the growth of prostate tumours developing within the bone.
The researchers found that their combination of non-invasive imaging techniques – which included magnetic resonance imaging (MRI) and single-photon emission computed tomography (SPECT) – produced the clearest images of tumour growth within bone to date, allowing them to accurately analyse the drug’s impact on the healthiness of the bones themselves.
The study, published in the Journal of the National Cancer Institute, found that in mice treated with cabozantinib, the drug showed signs of reducing tumour growth, preventing tumours from getting worse for 15 days. There was also evidence that the drug could provide some pain relief.
References
Graham T. J. et al., (2014) Preclinical evaluation of imaging biomarkers for prostate cancer bone metastasis and response to cabozantinib, Journal of the National Cancer Institute, 106(4):dju033. doi:10.1093/jnci/dju033
Discovering a link between lung cancer and a defect in the BRCA2 gene
A team at the ICR, led by Professor Richard Houlston, discovered a previously unknown link between lung cancer and a particular defect in the BRCA2 gene.
The defect, occurring in around 2% of the population, increases the risk of developing lung cancer in smokers by about 1.8 times.
The BRCA2 gene is best known for its role in breast cancer, but the large international study – published in the journal Nature Genetics – suggests an important link with lung cancer too.
Professor Houlston and colleagues at the ICR compared the DNA of more than 11,000 people with lung cancer and more than 15,000 people without the disease, looking for differences at specific points in their DNA. The study showed specific mutations to two genes, BRCA2 and CHEK2, significantly affect lung cancer risk in smokers.
When inherited, a particular defect in the BRCA2 gene called BRCA2 c.9976T was associated with a 1.8-fold increased risk of lung cancer. The link was particularly strong in patients with the most common lung cancer sub-type, called squamous cell lung cancer.
Smokers in general have a lifetime risk of developing lung cancer of around 15 per cent, far higher than in non-smokers. This study suggests that for smokers with BRCA2 mutations, around a quarter will develop lung cancer at some point in their lifetime.
In the future, patients with squamous cell lung cancer that carry this mutation could benefit from drugs specifically designed to be effective in cancers with BRCA mutations. A family of drugs called PARP inhibitors have shown success in clinical trials in patients with breast and ovarian cancer and BRCA mutations, although it is not known whether these could be effective in lung cancer.
References
Wang T. et al., (2014) Rare variants of large effect in BRCA2 and CHEK2 affect risk of lung cancer, Nature Genetics, 46, 736–741, doi:10.1038/ng.3002
Chemical tool can investigate effects of inhibiting key cancer protein
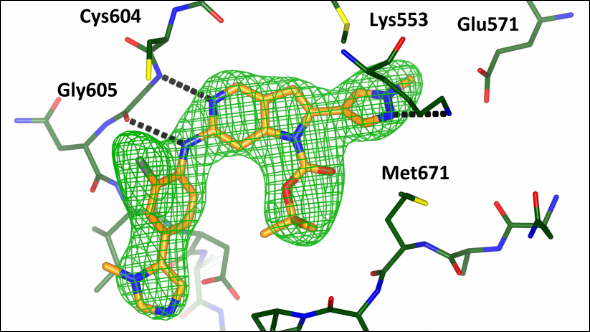
Scientists at the ICR have designed and synthesised a new molecule which will allow researchers to explore whether blocking a key cell division protein could be a possible therapeutic strategy in cancer.
The research, a collaboration between research groups led by Professor Julian Blagg, Dr Spiros Linardopoulos and Dr Rob van Montfort, could lead to the development of new drugs that target a protein called MPS1 – which has shown potential as a cancer drug target in early studies.
The new molecule – designed to bind with MPS1 as effectively as possible – fits precisely into a pocket within the MPS1 protein. Key to this work was the investigation into the shape of this pocket by protein X-ray crystallography in the laboratory of Dr Rob van Montfort.
MPS1, which is so important to life that it is present in humans in the same form as in yeast, helps all cells to assemble their DNA on cellular ‘rails’ during cell division. It is known to help drive the rapid division of several different cancer cell types, including breast, lung, prostate and bladder.
In their study, which was funded by Cancer Research UK, Breakthrough Breast Cancer and the Institute of Cancer Research, the researchers tested a range of molecules for their affinity to MPS1 – using X-ray crystal structures of the protein to design those that bound most strongly. They made new iterations of their molecules until they found one that bound more strongly and selectively to the MPS1 protein than other known inhibitors of MPS1.
Initial testing in mice transplanted with human tumour cells showed that an optimised molecule, called CCT251455, effectively inhibits MPS1.
In creating a new chemical tool for the study of MPS1, the study could help lead to new drugs that block cancer cell division.
References
Naud S et al, (2013). Structure-Based Design of Orally Bioavailable 1H-Pyrrolo[3,2-c]pyridine Inhibitors of Mitotic Kinase Monopolar Spindle 1 (MPS1). J Med Chem, 56(24): 10045–10065. doi:10.1021/jm401395s